This post is a write-up of the Twitter thread of the same topic, but long Twitter threads tend to get hard to read. So here goes:
During the day last Monday (2019-01-21), we noticed that the antiSMASH 5 results we were preparing for a course looked a bit odd for some of the multimodular NRPS and PKS clusters. As antiSMASH 5 is currently in beta, we initially suspected some antiSMASH bug and decided to investigate a bit further. Fortunately for us, it turned out that what we were seeing was not an antiSMASH issue but instead a problem with the NCBI RefSeq annotations with the current PGAP version.
Let me illustrate with an example. I will use the kirromycin gene cluster from Streptomyces collinus because that shows the problem and I happen to know it really well. I will provide screenshots of the antiSMASH results because while I did run all of this on the public antiSMASH web service, that deletes all job data after one month and I'm not sure how long this problem will hang around.
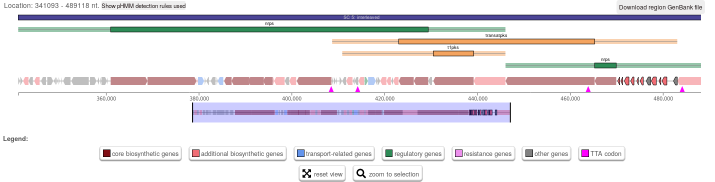
Basically the problem looks like this (click for a larger version):
Note the highlighted set of small ORFs on the right hand side of the cluster
(locus tags B446_RS38675, B446_RS38680, B446_RS38685, B446_RS38690,
B446_RS38695, B446_RS38700,B446_RS38705, B446_RS38710, B446_RS38715,
B446_RS38720, and B446_RS38725). This is how the results look for the
current record NC_021985.1 from RefSeq that I downloaded via the Entrez API
today. The date in the header line of the file is given as 10-NOV-2018.
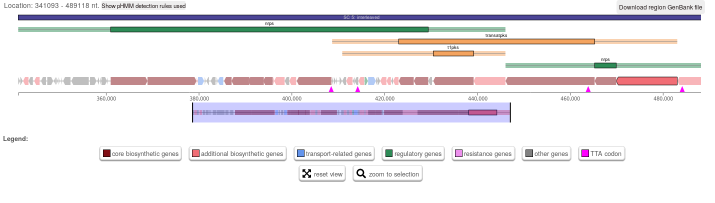
However, we would expect the cluster to look like this (click for a larger
version):
Note how instead the small ORFs in the previous image, the highlighted ORF with locus tag B446_RS37995 fills
the available space. This is another antiSMASH 5 run of today, but with a copy
of the NC_021985.1 record I downloaded early summer last year. The date in the
header line is given as 29-MAR-2018 for that one.
Now if we compare this to the manually curated kirromycin MIBiG entry that has the cluster going in the other direction, we see that the ORF that is broken in the current RefSeq annotation is the kirAII gene that contains a number of Trans-AT PKS modules and clearly is part of the gene cluster.
I will be reporting this issue to the NCBI RefSeq team after this post is live (so I can reference it in my issue report). Let's see how long it'll take to get this fixed.
Comments
comments powered by Disqus